Identification of the pseudoautosomal region of the X chromosome in sheep and sex prediction using the ARS-UI_Ramb_v2.0 genome assembly
DOI:
https://doi.org/10.31285/AGRO.29.1587Keywords:
ARS-I_Ramb_v2.0, pseudoautosomal region, sex prediction, X chromosomeAbstract
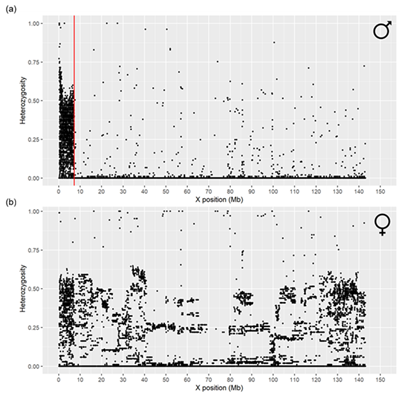
Sex determination through genotyping is an efficient tool for verifying recorded sex in sheep. Since males possess only one X chromosome, heterozygous genotypes should not be present in the non-pseudoautosomal region (nPAR) of the X chromosome. Therefore, determining the boundaries of the pseudoautosomal region (PAB) is necessary for a reliable sex prediction. Although it is recommended to use SNPs from both the X and Y chromosomes, the SNP chip we used did not contain Y chromosome markers. This study aimed to determine the pseudoautosomal region (PAR) on the X chromosome in sheep using the ARS-UI_Ramb_v2.0 genome assembly and test a method for sex prediction based solely on X chromosome SNPs. The training dataset was composed of 210 sheep from various breeds and crossbreeds, genotyped with the Ovine Infinium® HD SNP BeadChip to determine the PAR region. A validation dataset of 229 sheep was used to assess the accuracy of sex determination using the OvineSNP50 BeadChip. After quality control, the PAR region was found to span from 0 to 7.24 Mb and was identified by SNPs that exhibited high heterozygosity rates in males. The proposed approach, that uses only X chromosome SNPs, achieved a sex prediction accuracy of 94% with precision rates of 99% for males and 89% for females. This study showed that it is possible to predict sex in sheep using only X chromosome nPAR SNPs. This is the first study that has used this approach specifically using the ARS-UI_Ramb_v2.0 genome assembly.
Downloads
References
Aguilar I, Misztal I, Tsuruta S, Legarra A, Wang H. PREGSF90 – POSTGSF90: computational tools for the implementation of single-step genomic selection and genome-wide association with ungenotyped individuals in BLUPF90 programs. In: Proceedings 10th World Congress of Genetics Applied to Livestock Production. Vancouver: American Society of Animal Science; 2014. p. 680.
Baylis MS, Wayte DM, Owen JB. An XO/XX mosaic sheep with associated gonadal dysgenesis. Res Vet Sci. 1984;36(1):125-6. DOI: https://doi.org/10.1016/S0034-5288(18)32013-7
Berkowic Y, Edwards LA, Busch R, Pérez-Nogués M, Stevens SE, Booth M, McNabb BR. Male pseudohermaphrodite in a Shetland cross sheep: a case report. Clinical Theriogenology. 2017;9(4):605-13.
Berry DP, Spangler ML. Animal board invited review: practical applications of genomic information in livestock. Animal. 2023;17(11):100996. Doi: 10.1016/j.animal.2023.100996. DOI: https://doi.org/10.1016/j.animal.2023.100996
Bilton TP, Chappell AJ, Clarke SM, Brauning R, Dodds KG, McEwan JC, Rowe SJ. Using genotyping-by-sequencing to predict gender in animals. Anim Genet. 2019;50(3):307-10. Doi: 10.1111/age.12782. DOI: https://doi.org/10.1111/age.12782
Bruere AN, Marshall RB, Ward DP. Testicular hypoplasia and XXY sex chromosome complement in two rams: the ovine counterpart of Klinefelter's syndrome in man. J Reprod Fertil. 1969;19(1):103-8. Doi: 10.1530/jrf.0.0190103. DOI: https://doi.org/10.1530/jrf.0.0190103
Champely S. pwr: basic functions for power analysis [Internet]. Version 1.3-0. 2020 [cited 2025 May 27]. Available from: https://CRAN.R-project.org/package=pwr
Ciappesoni G, Marques C, Navajas EA, Peraza P, Carracelas B, Vera B, Van Lier E, De Barbieri I, Salada S, Monzalvo C, Castells D. Breeding for sheep parasite resistance in extensive production systems in Uruguay: from phenotype to genotype. In: Viljoen G, Garcia Podesta M, Boettcher P, editors. International Symposium on Sustainable Animal Production and Health - Current status and way forward. Rome: FAO; 2023. p. 224-36.
Johnson T, Keehan M, Harland C, Lopdell T, Spelman RJ, Davis SR, Rosen BD, Smith TPL, Couldrey C. Identification of the pseudoautosomal region in the Hereford bovine reference genome assembly ARS-UCD1.2. J Dairy Sci. 2019;102(4):3254-8. Doi: 10.3168/jds.2018-15638. DOI: https://doi.org/10.3168/jds.2018-15638
Katoh K, Rozewicki J, Yamada KD. MAFFT online service: multiple sequence alignment, interactive sequence choice and visualization. Brief Bioinform. 2019;20(4):1160-6. Doi: 10.1093/bib/bbx108. DOI: https://doi.org/10.1093/bib/bbx108
Kuhn M. Building Predictive Models in R Using the caret Package. J Stat Softw. 2008;28(5):1-26. Doi: 10.18637/jss.v028.i05 DOI: https://doi.org/10.18637/jss.v028.i05
Liu R, Low WY, Tearle R, Koren S, Ghurye J, Rhie A, Phillippy AM, Rosen BD, Bickhart DM, Smith TPL, Hiendleder S, Williams JL. New insights into mammalian sex chromosome structure and evolution using high-quality sequences from bovine X and Y chromosomes. BMC Genomics. 2019;20(1):1000. Doi: 10.1186/s12864-019-6364-z. DOI: https://doi.org/10.1186/s12864-019-6364-z
McClure MC, McCarthy J, Flynn P, McClure JC, Dair E, O'Connell DK, Kearney JF. SNP data quality control in a national beef and dairy cattle system and highly accurate SNP based parentage verification and identification. Front Genet. 2018;9:84. Doi: 10.3389/fgene.2018.00084. DOI: https://doi.org/10.3389/fgene.2018.00084
Medrano JF, Aasen E, Sharrow L. DNA extraction from nucleated red blood cells. Biotechniques. 1990;8(1):43.
Olagunju TA, Rosen BD, Neibergs HL, Becker GM, Davenport KM, Elsik CG, Hadfield TS, Koren S, Kuhn KL, Rhie A, Shira KA, Skibiel AL, Stegemiller MR, Thorne JW, Villamediana P, Cockett NE, Murdoch BM, Smith TPL. Telomere-to-telomere assemblies of cattle and sheep Y-chromosomes uncover divergent structure and gene content. Nat Commun. 2024;15(1):8277. Doi: 10.1038/s41467-024-52384-5. DOI: https://doi.org/10.1038/s41467-024-52384-5
Ramos Z, Garrick DJ, Blair HT, De Barbieri I, Ciappesoni G, Montossi F, Kenyon PR. Genetic trends for production and reproduction traits in ultrafine Merino sheep of Uruguay. J Anim Breed Genet. Forhcoming 2025. Doi: 10.1111/jbg.12937. DOI: https://doi.org/10.1111/jbg.12937
Raudsepp T, Chowdhary BP. Chromosome aberrations and fertility disorders in domestic animals. Annu Rev Anim Biosci. 2016;4:15-43. Doi: 10.1146/annurev-animal-021815-111239. DOI: https://doi.org/10.1146/annurev-animal-021815-111239
Raudsepp T, Das PJ, Avila F, Chowdhary BP. The pseudoautosomal region and sex chromosome aneuploidies in domestic species. Sex Dev. 2012;6(1-3):72-83. Doi: 10.1159/000330627. DOI: https://doi.org/10.1159/000330627
Ristanic M, Stanisic L, Maletic M, Glavinic U, Draskovic V, Aleksic N, Stanimirovic Z. Bovine foetal sex determination-Different DNA extraction and amplification approaches for efficient livestock production. Reprod Domest Anim. 2018;53(4):947-54. Doi: 10.1111/rda.13193. DOI: https://doi.org/10.1111/rda.13193
Ryan CA, Purfield DC, Matthews D, Canedo-Ribeiro C, Valldecabres A, Berry DP. Prevalence of sex-chromosome aneuploidy estimated using SNP genotype intensity information in a large population of juvenile dairy and beef cattle. J Anim Breed Genet. 2024;141(5):571-85. Doi: 10.1111/jbg.12866. DOI: https://doi.org/10.1111/jbg.12866
Shihabi M, Lukic B, Cubric-Curik V, Brajkovic V, Oršanić M, Ugarković D, Vostry L, Curik I. Identification of selection signals on the X-Chromosome in East Adriatic Sheep: a new complementary approach. Front Genet. 2022;13:887582. Doi: 10.3389/fgene.2022.887582. DOI: https://doi.org/10.3389/fgene.2022.887582
Wickham H. ggplot2: elegant graphics for data analysis [Internet]. [place unknown]: The tidyverse team; [cited 2025 May 27]. Available from: https://ggplot2.tidyverse.org
Zartman DL, Hinesley LL, Gnatkowski MW. A 53,X female sheep (Ovis aries). Cytogenet Cell Genet. 1981;30(1):54-8. Doi: 10.1159/000131589. DOI: https://doi.org/10.1159/000131589
Zhang I, Couldrey C, Sherlock R. Using genomic information to predict sex in dairy cattle. Proc N Z Soc Anim Prod. 2016;76:26-30.
Downloads
Published
How to Cite
Issue
Section
License
Copyright (c) 2025 Agrociencia Uruguay

This work is licensed under a Creative Commons Attribution 4.0 International License.
| Article metrics | |
|---|---|
| Abstract views | |
| Galley vies | |
| PDF Views | |
| HTML views | |
| Other views | |