Influence of under-vine soil management on bulk soil microbiota and properties in three Tannat vineyards in Uruguay
DOI:
https://doi.org/10.31285/AGRO.29.1698Keywords:
grapevine, permanent living mulch, herbicide, amplicon sequencing, soil microbiome, 16S rRNA, ITS2Abstract
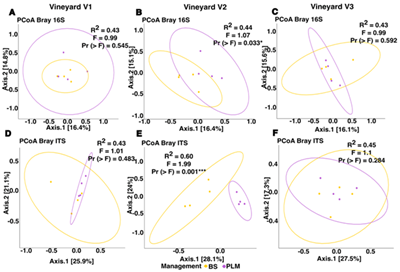
The vineyard soil microbiome plays a pivotal role in agroecosystem health and productivity, influenced by agricultural management and environmental factors. This study examined the composition of prokaryotic and fungal communities across three Tannat vineyards in Uruguay with the same conventional under-vine soil management (bare soil maintained with herbicides [BS]). Additionally, within each vineyard, a permanent living mulch (PLM) was established to explore the effects of this under-vine management on soil microbiota. The long-term cultivation of Tannat grapevines combined with consistent under-vine herbicide use may have contributed to a homogenization of soil microbial community composition across vineyards, despite differences in soil type, altitude, and management histories. A few differentially abundant taxa were detected: in Vineyard 3 Rubrobacter was less abundant compared to the other vineyards, while in Vineyard 1 the class Sordariomycetes and the genus Metarhizium were more abundant, and the genus Boeremia was less. Prokaryotic and fungal communities' composition analyses within vineyards revealed a significant impact of under-vine management solely in Vineyard 2, the one with a 10-year implementation of PLM. PLM improved soil properties such as basal respiration, soil protein, potentially oxidizable carbon and bulk density. Also, the family Latescibacteraceae, and the genera Cladophialophora, Nigrospora, and Pseudopithomyces were more abundant in PLM. Our findings emphasize the need for long-term studies to capture microbial responses to soil management. Future studies involving more sites and managements could identify deeper differences, aiding in the identification of viticultural zones based on microbial patterns.
Downloads
References
Alliaume F, Echeverria G, Ferrer M, González Barrios P. A study of the multivariate spatial variability of soil properties, and their association with vine vigor growing on a clayish soil. J Soil Sci Plant Nutr. 2024;24:3282-97. Doi: 10.1007/s42729-024-01751-8. DOI: https://doi.org/10.1007/s42729-024-01751-8
Andersen KS, Kirkegaard RH, Karst SM, Albertsen M. ampvis2: an R package to analyse and visualise 16S rRNA amplicon data. BioRxiv 299537 [Preprint]. 2018. Doi: 10.1101/299537. DOI: https://doi.org/10.1101/299537
Bacq-Labreuil A, Crawford J, Mooney SJ, Neal AL, Ritz K. Cover crop species have contrasting influence upon soil structural genesis and microbial community phenotype. Sci Rep. 2019;9(1):7473. Doi: 10.1038/s41598-019-43937-6. DOI: https://doi.org/10.1038/s41598-019-43937-6
Bais HP, Weir TL, Perry LG, Gilroy S, Vivanco JM. The role of root exudates in rhizosphere interactions with plants and other organisms. Annu Rev Plant Biol. 2006;57:233-66. Doi: 10.1146/annurev.arplant.57.032905.105159. DOI: https://doi.org/10.1146/annurev.arplant.57.032905.105159
Bates D, Mächler M, Bolker B, Walker S. Fitting linear mixed-effects models using lme4. J Stat Softw. 2015;67(1):1-48. Doi: 10.18637/jss.v067.i01. DOI: https://doi.org/10.18637/jss.v067.i01
Belda I, Zarraonaindia I, Perisin M, Palacios A, Acedo A. From vineyard soil to wine fermentation: microbiome approximations to explain the "terroir" concept. Front Microbiol. 2017;8:821. Doi: 10.3389/fmicb.2017.00821. DOI: https://doi.org/10.3389/fmicb.2017.00821
Berg G, Smalla K. Plant species and soil type cooperatively shape the structure and function of microbial communities in the rhizosphere. FEMS Microbiol Ecol. 2009;68(1):1-13. Doi: 10.1111/j.15746941.2009.00654.x. DOI: https://doi.org/10.1111/j.1574-6941.2009.00654.x
Bernaschina Y, Fresia P, Garaycochea S, Leoni C. Correction: permanent cover crop as a strategy to promote soil health and vineyard performance. Environmental Sustainability. 2023;6:295. Doi: 10.1007/s42398-023-00283-8. DOI: https://doi.org/10.1007/s42398-023-00283-8
Bettenfeld P, Cadena I Canals J, Jacquens L, Fernandez O, Fontaine F, van Schaik E, Courty PE, Trouvelot S. The microbiota of the grapevine holobiont: a key component of plant health. J Adv Res. 2022;40:1-15. Doi: 10.1016/j.jare.2021.12.008. DOI: https://doi.org/10.1016/j.jare.2021.12.008
Blanco‐Canqui H, Shaver TM, Lindquist JL, Shapiro CA, Elmore RW, Francis CA, Hergert GW. Cover crops and ecosystem services: insights from studies in temperate soils. Agron J. 2015;107(6):2449-74. Doi: 10.2134/agronj15.0086. DOI: https://doi.org/10.2134/agronj15.0086
Bokulich NA, Collins TS, Masarweh C, Allen G, Heymann H, Ebeler SE, Mills DA. Associations among wine grape microbiome, metabolome, and fermentation behavior suggest microbial contribution to regional wine characteristics. mBio. 2016;7(3):e00631-16. Doi: 10.1128/mBio.00631-16. DOI: https://doi.org/10.1128/mBio.00631-16
Burns KN, Kluepfel DA, Strauss SL, Bokulich NA, Cantu D, Steenwerth KL. Vineyard soil bacterial diversity and composition revealed by 16S rRNA genes: differentiation by geographic features. Soil Biol Biochem. 2015;91:232-47. Doi: 10.1016/j.soilbio.2015.09.002. DOI: https://doi.org/10.1016/j.soilbio.2015.09.002
Callahan BJ, McMurdie PJ, Rosen MJ, Han AW, Johnson AJ, Holmes SP. DADA2: high-resolution sample inference from Illumina amplicon data. Nat Methods. 2016;13(7):581-3. Doi: 10.1038/nmeth.3869. DOI: https://doi.org/10.1038/nmeth.3869
Chaparro JM, Sheflin AM, Manter DK, Vivanco JM. Manipulating the soil microbiome to increase soil health and plant fertility. Biol Fertil Soils. 2012;48(5):489-99. Doi: 10.1007/s00374-012-0691-4. DOI: https://doi.org/10.1007/s00374-012-0691-4
Chou MY, Vanden Heuvel J, Bell TH, Panke-Buisse K, Kao-Kniffin J. Vineyard under-vine floor management alters soil microbial composition, while the fruit microbiome shows no corresponding shifts. Sci Rep. 2018;8(1):11039. Doi: 10.1038/s41598-018-29346-1. DOI: https://doi.org/10.1038/s41598-018-29346-1
Coller E, Cestaro A, Zanzotti R, Bertoldi D, Pindo M, Larger S, Albanese D, Mescalchin E, Donati C. Microbiome of vineyard soils is shaped by geography and management. Microbiome. 2019;7(1):140. Doi: 10.1186/s40168-019-0758-7. DOI: https://doi.org/10.1186/s40168-019-0758-7
Coniberti A, Ferrari V, Disegna E, Dellacassa E, Lakso AN. Under-trellis cover crop and deficit irrigation to regulate water availability and enhance Tannat wine sensory attributes in a humid climate. Sci Hortic. 2018;235:244-52. Doi: 10.1016/j.scienta.2018.03.018. DOI: https://doi.org/10.1016/j.scienta.2018.03.018
Coniberti A, Ferrari V, Disegna E, García Petillo M, Lakso AN. Complete vineyard floor cover crop to reduce grapevine susceptibility to bunch rot. Eur J Agron. 2018;99:167-76. Doi: 10.1016/j.eja.2018.07.006. DOI: https://doi.org/10.1016/j.eja.2018.07.006
Eichhorn KW, Lorenz DH. Phaenologische entwicklungsstadien der rebe. Nachr Dtsch Pflanzenschutzd. 1977;29:119-20.
Ferrer M, Pedocchi R, Michelazzo M, González-Neves G, Carbonneau A. Delimitación y descripción de regiones vitícolas del Uruguay en base al método de clasificación climática multicriterio utilizando índices bioclimáticos adaptados a las condiciones del cultivo. Agrocienc Urug. 2007;11(1):47-56. Doi: 10.31285/AGRO.11.768. DOI: https://doi.org/10.31285/AGRO.11.768
Fuchs B, Saikkonen K, Damerau A, Yang B, Helander M. Herbicide residues in soil decrease microbe-mediated plant protection. Plant Biol (Stuttg). 2023;25(4):571-8. Doi: 10.1111/plb.13517. DOI: https://doi.org/10.1111/plb.13517
Gobbi A, Acedo A, Imam N, Santini RG, Ortiz-Álvarez R, Ellegaard-Jensen L, Belda I, Hansen LH. A global microbiome survey of vineyard soils highlights the microbial dimension of viticultural terroirs. Commun Biol. 2022;5(1):241. Doi: 10.1038/s42003-022-03202-5. DOI: https://doi.org/10.1038/s42003-022-03202-5
Hartmann M, Six J. Soil structure and microbiome functions in agroecosystems. Nat Rev Earth Environ. 2023;4:4-18. Doi: 10.1038/s43017-022-00366-w. DOI: https://doi.org/10.1038/s43017-022-00366-w
Hendgen M, Hoppe B, Döring J, Friedel M, Kauer R, Frisch M, Dahl A, Kellner H. Effects of different management regimes on microbial biodiversity in vineyard soils. Sci Rep. 2018;8(1):9393. Doi: 10.1038/s41598-018-27743-0. DOI: https://doi.org/10.1038/s41598-018-27743-0
Hothorn T, Bretz F, Westfall P. Simultaneous inference in general parametric models. Biom J. 2008;50(3):346-63. Doi: 10.1002/bimj.200810425. DOI: https://doi.org/10.1002/bimj.200810425
INAVI. Reporte anual: registro de viñedos [Internet]. Montevideo: INAVI; 2024 [cited 2025 Sep 29]. 35p. Available from: https://www.inavi.com.uy/uploads/vinedo/346bb91b94f57f26d3e293b52fc0059a83b273cf.pdf
Knight S, Klaere S, Fedrizzi B, Goddard MR. Regional microbial signatures positively correlate with differential wine phenotypes: evidence for a microbial aspect to terroir. Sci Rep. 2015;5:14233. Doi: 10.1038/srep14233. DOI: https://doi.org/10.1038/srep14233
Kuznetsova A, Brockhoff PB, Christensen RHB. lmertest package: tests in linear mixed effects models. J Stat Softw. 2017;82(13):1-26. Doi: 10.18637/jss.v082.i13. DOI: https://doi.org/10.18637/jss.v082.i13
Lahti L, Sudarshan S. microbiome: tools for microbiome analysis in R [Internet]. Version 1.0.2. 2017 [cited 2025 Sep 29]. Available from: https://bioconductor.org/packages/microbiome
Lenth RV. emmeans: Estimated Marginal Means, aka Least-Squares Means [Internet]. Version 1.10.3-090006. 2024 [cited 2025 Sep 29]. Available from: https://CRAN.R-project.org/package=emmeans
Lin H, Peddada SD. Analysis of compositions of microbiomes with bias correction. Nat Commun. 2020;11(1):3514. Doi: 10.1038/s41467-020-17041-7. DOI: https://doi.org/10.1038/s41467-020-17041-7
Longa CMO, Nicola L, Antonielli L, Mescalchin E, Zanzotti R, Turco E, Pertot I. Soil microbiota respond to green manure in organic vineyards. J Appl Microbiol. 2017;123(6):1547-60. Doi: 10.1111/jam.13606. DOI: https://doi.org/10.1111/jam.13606
Longepierre M, Widmer F, Keller T, Weisskopf P, Colombi T, Six J, Hartmann M. Limited resilience of the soil microbiome to mechanical compaction within four growing seasons of agricultural management. ISME Commun. 2021;1(1):44. Doi: 10.1038/s43705-021-00046-8. DOI: https://doi.org/10.1038/s43705-021-00046-8
Lüdecke D, Ben-Shachar M, Patil I, Waggoner P, Makowski D. Performance: an R Package for assessment, comparison and testing of statistical models. J Open Source Softw. 2021;6(60):3139. Doi: 10.21105/joss.03139. DOI: https://doi.org/10.21105/joss.03139
McKenzie N, Jacquier D, Isbell R, Brown K. Australian soils and landscapes: an illustrated compendium. Collingwood: CSIRO; 2004. 416p. DOI: https://doi.org/10.1071/9780643100732
McMurdie PJ, Holmes S. phyloseq: an R package for reproducible interactive analysis and graphics of microbiome census data. PLoS One. 2013;8(4):e61217. Doi: 10.1371/journal.pone.0061217. DOI: https://doi.org/10.1371/journal.pone.0061217
Mocali S, Kuramae EE, Kowalchuk GA, Fornasier F, Priori S. Microbial functional diversity in vineyard soils: sulfur metabolism and links with grapevine plants and wine quality. Front Environ Sci. 2020;8:75. Doi: 10.3389/fenvs.2020.00075 DOI: https://doi.org/10.3389/fenvs.2020.00075
Moebius-Clune BN, Moebius-Clune DJ, Gigino BK, Idowu OJ, Schindelbeck RR, Ristow AJ, van Es HM, Thies JE, Shayler HA, McBride MB, Kurtz KSM, Wolfe DW, Abawi GS. Comprehensive assessment of soil health: the Cornell framework manual. 3rd ed. Ithaca: Cornell University; 2016. 123p.
Molfino JH. Estimación del agua disponible en los grupos CONEAT: metodología empleada [Internet]. 2009 [cited 2025 Sep 29]. Available from: https://www.gub.uy/ministerio-ganaderia-agricultura-pesca/comunicacion/publicaciones/estimacion-agua-disponible-grupos-coneat-metodologia-empleada
Nilsson RH, Larsson KH, Taylor AFS, Bengtsson-Palme J, Jeppesen TS, Schigel D, Kennedy P, Picard K, Glöckner FO, Tedersoo L, Saar I, Kõljalg U, Abarenkov K. The UNITE database for molecular identification of fungi: handling dark taxa and parallel taxonomic classifications. Nucleic Acids Res. 2019;47(D1):D259-D264. Doi: 10.1093/nar/gky1022. DOI: https://doi.org/10.1093/nar/gky1022
Oksanen J, Simpson GL, Blanchet FG, Kindt R, Legendre P, Minchin PR, O’Hara RB, Solymos P, Stevens MHH, Wagner H, Barbour M, Bedward M, Bolker B, Borcard D, Carvalho G, Chirico M, Caceres MD, Durand S, Evangelista HBA, FitzJohn R. vegan: Community Ecology Package [Internet]. Version 2.5-7. 2020 [cited 2025 Sep 29]. Available from: https://CRAN.R-project.org/package=vegan
Peng Z, Qian X, Liu Y, Li X, Gao H, An Y, Qi J, Jiang L, Zhang Y, Chen S, Pan H, Chen B, Liang C, van der Heijden MGA, Wei G, Jiao S. Land conversion to agriculture induces taxonomic homogenization of soil microbial communities globally. Nat Commun. 2024;15(1):3624. Doi: 10.1038/s41467-024-47348-8. DOI: https://doi.org/10.1038/s41467-024-47348-8
Pingel M, Reineke A, Leyer I. Disentangling the mixed effects of soil management on microbial diversity and soil functions: a case study in vineyards. Sci Rep. 2023;13(1):3568. Doi: 10.1038/s41598-023-30338-z. DOI: https://doi.org/10.1038/s41598-023-30338-z
Pini F, Galardini M, Bazzicalupo M, Mengoni A. Plant-bacteria association and symbiosis: are there common genomic traits in alphaproteobacteria? Genes (Basel). 2011;2(4):1017-32. Doi: 10.3390/genes2041017. DOI: https://doi.org/10.3390/genes2041017
Quast C, Pruesse E, Yilmaz P, Gerken J, Schweer T, Yarza P, Peplies J, Glöckner FO. The SILVA ribosomal RNA gene database project: improved data processing and web-based tools. Nucleic Acids Res. 2013;41(Database issue):D590-6. Doi: 10.1093/nar/gks1219. DOI: https://doi.org/10.1093/nar/gks1219
Schmidt PA, Bálint M, Greshake B, Bandow C, Römbke J, Schmitt I. Illumina metabarcoding of a soil fungal community. Soil Biol Biochem. 2013;65:128-32. Doi: 10.1016/j.soilbio.2013.05.014. DOI: https://doi.org/10.1016/j.soilbio.2013.05.014
Sharma P, Singh A, Kahlon CS, Brar AS, Grover KK, Dia M, Steiner RL. The role of cover crops towards sustainable soil health and agriculture: a review paper. Am J Plant Sci. 2018;9(9):1935-51. Doi: 10.4236/ajps.2018.99140. DOI: https://doi.org/10.4236/ajps.2018.99140
Takahashi S, Tomita J, Nishioka K, Hisada T, Nishijima M. Development of a prokaryotic universal primer for simultaneous analysis of Bacteria and Archaea using next-generation sequencing. PLoS One. 2014;9(8):e105592. Doi: 10.1371/journal.pone.0105592. DOI: https://doi.org/10.1371/journal.pone.0105592
Tkacz A, Cheema J, Chandra G, Grant A, Poole PS. Stability and succession of the rhizosphere microbiota depends upon plant type and soil composition. ISME J. 2015;9(11):2349-59. Doi: 10.1038/ismej.2015.41. DOI: https://doi.org/10.1038/ismej.2015.41
Tsoy OV, Ravcheev DA, Čuklina J, Gelfand MS. Nitrogen fixation and molecular oxygen: comparative genomic reconstruction of transcription regulation in alphaproteobacteria. Front Microbiol. 2016;7:1343. Doi: 10.3389/fmicb.2016.01343. DOI: https://doi.org/10.3389/fmicb.2016.01343
United States Department of Agriculture, NRCS. Soil taxonomy: a basic system of soil classification for making and interpreting soil surveys [Internet]. 2nd ed. Washington: USDA; 1999 [cited 2025 Sep 29]. 886p. Available from: https://www.nrcs.usda.gov/resources/guides-and-
instructions/soil-taxonomy
Vanden Heuvel J, Centinari M. Under-vine vegetation mitigates the impacts of excessive precipitation in vineyards. Front Plant Sci. 2021;12:713135. Doi: 10.3389/fpls.2021.713135. DOI: https://doi.org/10.3389/fpls.2021.713135
Wardle DA, Bardgett RD, Klironomos JN, Setälä H, van der Putten WH, Wall DH. Ecological linkages between aboveground and belowground biota. Science. 2004;304(5677):1629-33. Doi: 10.1126/science.1094875. DOI: https://doi.org/10.1126/science.1094875
Wickham H, Chang W, Pedersen TL, Wilke CO, Woo K, Yutani H, Bryan J, Csárdi G, Grolemund G, Hayes A, Henry L, Hester J, Kuhn M, Lin Pedersen T, Miller E, Milton Bache S, Müller K, Ooms J, Robinson D, Seidel D. ggplot2: Create elegant data visualisations using the grammar of graphics [Internet]. Version 3.4.0. 2023 [cited 2025 Sep 29]. Available from: https://CRAN.R-project.org/package=ggplot2
Yan H, Ge C, Zhou J, Li J. Diversity of soil fungi in the vineyards of Changli region in China. Can J Microbiol. 2022;68(5):341-52. Doi: 10.1139/cjm-2021-0337. DOI: https://doi.org/10.1139/cjm-2021-0337
Zarraonaindia I, Owens SM, Weisenhorn P, West K, Hampton-Marcell J, Lax S, Bokulich NA, Mills DA, Martin G, Taghavi S, van der Lelie D, Gilbert JA. The soil microbiome influences grapevine-associated microbiota. mBio. 2015;6(2):e02527-14. Doi: 10.1128/mBio.02527-14. DOI: https://doi.org/10.1128/mBio.02527-14
Downloads
Published
How to Cite
Issue
Section
License
Copyright (c) 2025 Agrociencia Uruguay

This work is licensed under a Creative Commons Attribution 4.0 International License.
| Article metrics | |
|---|---|
| Abstract views | |
| Galley vies | |
| PDF Views | |
| HTML views | |
| Other views | |