Identificação e caracterização de peptídeos antimicrobianos no Transcriptoma de Feijoa sellowiana
DOI:
https://doi.org/10.31285/AGRO.29.1556Palavras-chave:
AMPs, goiabeira-serrana, validação gênica, transcriptômica de novo, defensinasResumo
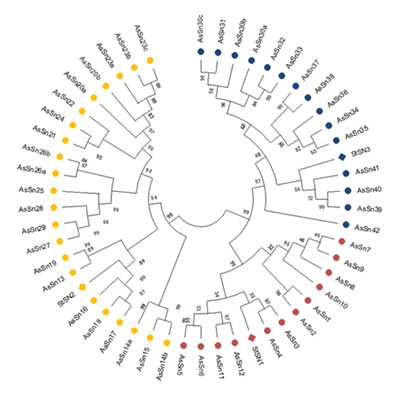
Os peptídeos antimicrobianos (AMPs) são pequenos polipeptídios presentes em uma grande diversidade de organismos filogeneticamente distantes, incluindo plantas. Eles fazem parte da resposta imunológica inata, permitindo uma ação rápida com um custo energético menor do que o sistema imunológico adaptativo. Diversas aplicações desses peptídeos como agentes anti-infecciosos foram demonstradas. Apesar de seu significativo potencial, a bioprospecção de AMPs na flora nativa do Uruguai ainda é incipiente. O objetivo deste estudo foi identificar e caracterizar genes que codificam seis famílias de peptídeos antimicrobianos compreendendo: defensinas, snakins, tioninas, hevein-like, proteínas de transferência de lipídios (LTPs) e ciclotídeos, na espécie frutífera nativa Feijoa sellowiana (Myrtaceae), comumente conhecida como goiabeira-serrana. A busca foi realizada usando BLAST, baseada em um transcriptoma de novo de folha e flor, utilizando seqüências de referência. As relações entre F. sellowiana e sequências de proteínas de referência foram caracterizadas por meio de alinhamentos múltiplos e análise filogenética. A estrutura exon-íntron e isoformas de AMPs de F. sellowiana foram analisadas por amplificação por PCR em DNA genômico. Este estudo identificou 23 defensinas, 49 snakins, 7 tioninas, 12 hevein-like, 87 LTPs; nenhuma evidência de transcrições de ciclotídeo foi encontrada. A estrutura exon-íntron esperada foi confirmada para cinco seqüências, pertencentes às famílias defensinas, esnaquinas e hevein-like.
Downloads
Referências
Almaghrabi B, Ali MA, Zahoor A, Shah KH, Bohlmann H. Arabidopsis thionin-like genes are involved in resistance against the beet-cyst nematode (Heterodera schachtii). Plant Physiol Biochem. 2019;140:55-67. Doi: 10.1016/j.plaphy.2019.05.005. DOI: https://doi.org/10.1016/j.plaphy.2019.05.005
Apweiler R, Bairoch A, Wu CH, Barker WC, Boeckmann B, Ferro S, Gasteiger E, Huang H, Lopez R, Magrane M, Martin MJ, Natale DA, O'Donovan C, Redaschi N, Yeh LS. UniProt: the Universal Protein knowledgebase. Nucleic Acids Res. 2004;32(Database issue):D115-9. Doi: 10.1093/nar/gkh131. DOI: https://doi.org/10.1093/nar/gkh131
Benko-Iseppon AM, Galdino SL, Calsa T Jr, Kido EA, Tossi A, Belarmino LC, Crovella S. Overview on plant antimicrobial peptides. Curr Protein Pept Sci. 2010;11(3):181-8. Doi: 10.2174/138920310791112075. DOI: https://doi.org/10.2174/138920310791112075
Berman HM, Westbrook J, Feng Z, Gilliland G, Bhat TN, Weissig H, Shindyalov IN, Bourne PE. The Protein Data Bank. Nucleic Acids Res. 2000;28(1):235-42. Doi: 10.1093/nar/28.1.235. DOI: https://doi.org/10.1093/nar/28.1.235
Berrocal-Lobo M, Segura A, Moreno M, López G, García-Olmedo F, Molina A. Snakin-2, an antimicrobial peptide from potato whose gene is locally induced by wounding and responds to pathogen infection. Plant Physiol. 2002;128(3):951-61. Doi: 10.1104/pp.010685. DOI: https://doi.org/10.1104/pp.010685
Bohlmann H, Broekaert W. The role of Thionins in plant protection. Crit Rev Plant Sci. 1994;13(1):1-16. Doi: 10.1080/07352689409701905. DOI: https://doi.org/10.1080/07352689409701905
Brogden KA. Antimicrobial peptides: pore formers or metabolic inhibitors in bacteria? Nat Rev Microbiol. 2005;3(3):238-50. Doi: 10.1038/nrmicro1098. DOI: https://doi.org/10.1038/nrmicro1098
Carvalho AO, Gomes VM. Plant defensins--prospects for the biological functions and biotechnological properties. Peptides. 2009;30(5):1007-20. Doi: 10.1016/j.peptides.2009.01.018. DOI: https://doi.org/10.1016/j.peptides.2009.01.018
Castro MS, Gerhardt IR, Orrù S, Pucci P, Bloch C Jr. Purification and characterization of a small (7.3 kDa) putative lipid transfer protein from maize seeds. J Chromatogr B Analyt Technol Biomed Life Sci. 2003;794(1):109-14. Doi: 10.1016/s1570-0232(03)00423-9. DOI: https://doi.org/10.1016/S1570-0232(03)00423-9
Craik DJ, Daly NL, Bond T, Waine C. Plant cyclotides: a unique family of cyclic and knotted proteins that defines the cyclic cystine knot structural motif. J Mol Biol. 1999;294(5):1327-36. Doi: 10.1006/jmbi.1999.3383. DOI: https://doi.org/10.1006/jmbi.1999.3383
Doyle J. DNA protocols for plants-CTAB total DNA isolation. In: Hewitt GM, Johnston AWB, Young JPW, editors. Molecular techniques in taxonomy. Berlin: Springer; 1991. p. 283-93. Doi: 10.1007/978-3-642-83962-7_18. DOI: https://doi.org/10.1007/978-3-642-83962-7_18
Ganz T. Defensins: antimicrobial peptides of vertebrates. C R Biol. 2004;327(6):539-49. Doi: 10.1016/j.crvi.2003.12.007. DOI: https://doi.org/10.1016/j.crvi.2003.12.007
Goodstein DM, Shu S, Howson R, Neupane R, Hayes RD, Fazo J, Mitros T, Dirks W, Hellsten U, Putnam N, Rokhsar DS. Phytozome: a comparative platform for green plant genomics. Nucleic Acids Res. 2012;40(Database issue):D1178-86. Doi: 10.1093/nar/gkr944. DOI: https://doi.org/10.1093/nar/gkr944
Hall TA. BioEdit: a user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucl Acids Symp Ser. 1999;41:95-8.
Joint Genome Institute. Phytozome [Internet]. Berkeley: University of California; 2016-2025 [cited 2025 May 27]. Available from: https://phytozome-next.jgi.doe.gov/
Kaur J, Sagarman US, Shah D. Can plant defensins be used to engineer durable commercially useful fungal resistance in crop plants? Fungal Biol Rev. 2011;25(3):128-35. Doi: 10.1016/j.fbr.2011.07.004. DOI: https://doi.org/10.1016/j.fbr.2011.07.004
Larrañaga P, Diaz-Dellavalle P, Cabrera A, Alem D, Leoni C, Almeida-Souza A, Giovanni-De-Simone S, Dalla-Rizza M. Activity of naturally derived antimicrobial peptides against filamentous fungi relevant for agriculture. Sustain Agric Res. 2012;1(2):211-21. DOI: https://doi.org/10.5539/sar.v1n2p211
Lay FT, Anderson MA. Defensins--components of the innate immune system in plants. Curr Protein Pept Sci. 2005;6(1):85-101. Doi: 10.2174/1389203053027575. DOI: https://doi.org/10.2174/1389203053027575
Li Z, Gao J, Wang G, Wang S, Chen K, Pu W, Wang Y, Xia Q, Fan X. Genome-Wide Identification and Characterization of GASA Gene Family in Nicotiana tabacum. Front Genet. 2022;12:768942. Doi: 10.3389/fgene.2021.768942. DOI: https://doi.org/10.3389/fgene.2021.768942
Nahirñak V, Almasia NI, Hopp HE, Vazquez-Rovere C. Snakin/GASA proteins: involvement in hormone crosstalk and redox homeostasis. Plant Signal Behav. 2012;7(8):1004-8. Doi: 10.4161/psb.20813. DOI: https://doi.org/10.4161/psb.20813
Nahirñak V, Rivarola M, Gonzalez de Urreta M, Paniego N, Hopp H, Almasia N, Vazquez-Rovere C. Genome-wide analysis of the Snakin/GASA gene family in Solanum tuberosum cv. Kennebec. Am J Potato Res. 2016;93(2):172-88. Doi: 10.1007/s12230-016-9494-8. DOI: https://doi.org/10.1007/s12230-016-9494-8
Nawrot R, Barylski J, Nowicki G, Broniarczyk J, Buchwald W, Goździcka-Józefiak A. Plant antimicrobial peptides. Folia Microbiol (Praha). 2014;59(3):181-96. Doi: 10.1007/s12223-013-0280-4. DOI: https://doi.org/10.1007/s12223-013-0280-4
Oberti H, Gutierrez-Gonzalez J, Pritsch C. A first de novo transcriptome assembly of feijoa (Acca sellowiana [Berg] Burret) reveals key genes involved in flavonoid biosynthesis. Plant Genome. 2024;17(3):e20501. Doi: 10.1002/tpg2.20501. DOI: https://doi.org/10.1002/tpg2.20501
Pestana-Calsa MC, Ribeiro IL, Calsa T Jr. Bioinformatics-coupled molecular approaches for unravelling potential antimicrobial peptides coding genes in Brazilian native and crop plant species. Curr Protein Pept Sci. 2010;11(3):199-209. Doi: 10.2174/138920310791112138. DOI: https://doi.org/10.2174/138920310791112138
Pujade-Renaud V, Sanier C, Cambillau L, Pappusamy A, Jones H, Ruengsri N, Tharreau D, Chrestin H, Montoro P, Narangajavana J. Molecular characterization of new members of the Hevea brasiliensis hevein multigene family and analysis of their promoter region in rice. Biochim Biophys Acta. 2005;1727(3):151-61. Doi: 10.1016/j.bbaexp.2004.12.013. DOI: https://doi.org/10.1016/j.bbaexp.2004.12.013
Rivas M, Puppo M, Baccino E, Quezada M, Franco J, Pritsch C. Phenotypic and molecular diversity of wild populations of Acca sellowiana (Berg.) burret in the Southern Area of natural distribution. Horticulturae. 2024;10(4):360. Doi: 10.3390/horticulturae10040360. DOI: https://doi.org/10.3390/horticulturae10040360
Rizwan Z, Aslam N, Zafar F, Humma R, Jamil A. Isolation of novel cyclotide encoding genes from some Solanaceae species and evolutionary link to other families. Pak J Agri Sci. 2021;58(1):169-77. Doi: 10.21162/PAKJAS/21.789.
Rodríguez Decuadro S. Prospección de defensinas y esnaquinas de plantas nativas para el desarrollo de nuevos agentes antimicrobianos [doctoral dissertation]. Montevideo (UY): Universidad de la República, Facultad de Ciencias; 2018. 122p.
Rodríguez-Decuadro S, Barraco-Vega M, Dans PD, Pandolfi V, Benko-Iseppon AM, Cecchetto G. Antimicrobial and structural insights of a new snakin-like peptide isolated from Peltophorum dubium (Fabaceae). Amino Acids. 2018;50(9):1245-59. Doi: 10.1007/s00726-018-2598-3. DOI: https://doi.org/10.1007/s00726-018-2598-3
Rodríguez-Decuadro S, da Rosa G, Radío S, Barraco-Vega M, Benko-Iseppon AM, Dans PD, Smircich P, Cecchetto G. Antimicrobial peptides in the seedling transcriptome of the tree legume Peltophorum dubium. Biochimie. 2021;180:229-42. Doi: 10.1016/j.biochi.2020.11.005. DOI: https://doi.org/10.1016/j.biochi.2020.11.005
Rodríguez-Decuadro S, Dans PD, Borba MA, Benko-Iseppon AM, Cecchetto G. Gene isolation and structural characterization of a legume tree defensin with a broad spectrum of antimicrobial activity. Planta. 2019;250(5):1757-72. Doi: 10.1007/s00425-019-03260-w. DOI: https://doi.org/10.1007/s00425-019-03260-w
Santos-Silva CAD, Ferreira-Neto JRC, Amador VC, Bezerra-Neto JP, Vilela LMB, Binneck E, Rêgo MS, da Silva MD, Mangueira de Melo ALT, da Silva RH, Benko-Iseppon AM. From gene to transcript and peptide: a deep overview on non-specific Lipid Transfer Proteins (nsLTPs). Antibiotics (Basel). 2023;12(5):939. Doi: 10.3390/antibiotics12050939. DOI: https://doi.org/10.3390/antibiotics12050939
Sayers EW, Beck J, Bolton EE, Brister JR, Chan J, Connor R, Feldgarden M, Fine AM, Funk K, Hoffman J, Kannan S, Kelly C, Klimke W, Kim S, Lathrop S, Marchler-Bauer A, Murphy TD, O'Sullivan C, Schmieder E, Skripchenko Y, Stine A, Thibaud-Nissen F, Wang J, Ye J, Zellers E, Schneider VA, Pruitt KD. Database resources of the National Center for Biotechnology Information in 2025. Nucleic Acids Res. 2025;53(D1):D20-D29. Doi: 10.1093/nar/gkae979. DOI: https://doi.org/10.1093/nar/gkae979
Segura A, Moreno M, Madueño F, Molina A, García-Olmedo F. Snakin-1, a peptide from potato that is active against plant pathogens. Mol Plant Microbe Interact. 1999;12(1):16-23. Doi: 10.1094/MPMI.1999.12.1.16. DOI: https://doi.org/10.1094/MPMI.1999.12.1.16
Selitrennikoff CP. Antifungal proteins. Appl Environ Microbiol. 2001;67(7):2883-94. Doi: 10.1128/AEM.67.7.2883-2894.2001. DOI: https://doi.org/10.1128/AEM.67.7.2883-2894.2001
Shang C, Ye T, Zhou Q, Chen P, Li X, Li W, Chen S, Hu Z, Zhang W. Genome-wide identification and bioinformatics analyses of host defense peptides Snakin/GASA in mangrove plants. Genes (Basel). 2023;14(4):923. Doi: 10.3390/genes14040923. DOI: https://doi.org/10.3390/genes14040923
Slavokhotova AA, Shelenkov AA, Korostyleva TV, Rogozhin EA, Melnikova NV, Kudryavtseva AV, Odintsova TI. Defense peptide repertoire of Stellaria media predicted by high throughput next generation sequencing. Biochimie. 2017;135:15-27. Doi: 10.1016/j.biochi.2016.12.017. DOI: https://doi.org/10.1016/j.biochi.2016.12.017
Slavokhotova AA, Shelenkov AA, Odintsova TI. Prediction of Leymus arenarius (L.) antimicrobial peptides based on de novo transcriptome assembly. Plant Mol Biol. 2015;89(3):203-14. Doi: 10.1007/s11103-015-0346-6. DOI: https://doi.org/10.1007/s11103-015-0346-6
Sun B, Zhao X, Gao J, Li J, Xin Y, Zhao Y, Liu Z, Feng H, Tan C. Genome-wide identification and expression analysis of the GASA gene family in Chinese cabbage (Brassica rapa L. ssp. pekinensis). BMC Genomics. 2023;24(1):668. Doi: 10.1186/s12864-023-09773-9. DOI: https://doi.org/10.1186/s12864-023-09773-9
Tamura K, Peterson D, Peterson N, Stecher G, Nei M, Kumar S. MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol Biol Evol. 2011;28(10):2731-9. Doi: 10.1093/molbev/msr121. DOI: https://doi.org/10.1093/molbev/msr121
Van Damme EJ, Charels D, Roy S, Tierens K, Barre A, Martins JC, Rougé P, Van Leuven F, Does M, Peumans WJ. A gene encoding a hevein-like protein from elderberry fruits is homologous to PR-4 and class V chitinase genes. Plant Physiol. 1999;119(4):1547-56. Doi: 10.1104/pp.119.4.1547. DOI: https://doi.org/10.1104/pp.119.4.1547
Van Parijs J, Broekaert WF, Goldstein IJ, Peumans WJ. Hevein: an antifungal protein from rubber-tree (Hevea brasiliensis) latex. Planta. 1991;183(2):258-64. Doi: 10.1007/BF00197797. DOI: https://doi.org/10.1007/BF00197797
Yeaman MR, Yount NY. Mechanisms of antimicrobial peptide action and resistance. Pharmacol Rev. 2003;55(1):27-55. Doi: 10.1124/pr.55.1.2. DOI: https://doi.org/10.1124/pr.55.1.2
Zasloff M. Antimicrobial peptides of multicellular organisms. Nature. 2002;415(6870):389-95. Doi: 10.1038/415389a. DOI: https://doi.org/10.1038/415389a
Zhu F. Chemical and biological properties of feijoa (Acca sellowiana). Trends Food Sci Technol. 2018;81:121-31. Doi: 10.1016/j.tifs.2018.09.008. DOI: https://doi.org/10.1016/j.tifs.2018.09.008
Downloads
Publicado
Como Citar
Edição
Seção
Licença
Copyright (c) 2025 Agrociencia Uruguay

Este trabalho está licenciado sob uma licença Creative Commons Attribution 4.0 International License.
| Métricas do artigo | |
|---|---|
| Vistas abstratas | |
| Visualizações da cozinha | |
| Visualizações de PDF | |
| Visualizações em HTML | |
| Outras visualizações | |