Regulação epigenética e memórias
possíveis aplicações à videira no Uruguai
DOI:
https://doi.org/10.31285/AGRO.28.1267Palavras-chave:
epigenética, metilação do ADN, modificações das histonas, videira, UruguaiResumo
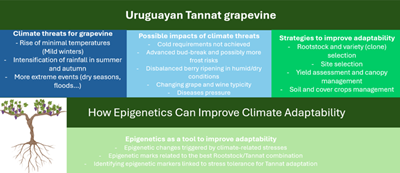
Este artigo explora o conhecimento atual da regulação epigenética nas videiras, realçando a sua importância em plantas propagadas clonalmente com diversidade genética limitada, como as videiras. Os principais processos epigenéticos, incluindo a metilação do ADN e as modificações das histonas, moldam a estrutura da cromatina, influenciando a expressão genética. O metiloma da folha da videira revela semelhanças com o de outras plantas propagadas clonalmente, salientando baixos níveis de metilação em contextos específicos. A regulação epigenética contribui para a plasticidade fenotípica da videira, a diversidade clonal e um diálogo intrigante entre parceiros enxertados. Estes mecanismos constituem uma parte vital da memória das plantas, especialmente face às alterações climáticas. No Uruguai, onde a indústria do vinho enfrenta desafios climáticos, a Tannat é uma variedade emblemática adaptada aos nossos sistemas de produção ambiental. No entanto, as previsões de alterações climáticas na região incluem o aumento das temperaturas, a alteração dos padrões de precipitação e o aumento dos fenômenos extremos, o que poderia afetar alguns aspectos da sua adaptação (rendimentos, qualidade dos bagos e tipicidade, entre outros). As estratégias de gestão da vinha, juntamente com o melhoramento vegetal, são essenciais para a adaptação. O artigo apela ao desenvolvimento urgente de estratégias inovadoras que utilizem variações epigenéticas hereditárias, apresentando uma abordagem mais rápida e eficiente para o melhoramento da videira para tolerância ao stress na era das alterações climáticas.
Downloads
Referências
Anastasiadi D, Venney CJ, Bernatchez L, Wellenreuther M. Epigenetic inheritance and reproductive mode in plants and animals. Trends Ecol Evol. 2021;36(12):1124-40. Doi: 10.1016/j.tree.2021.08.006. DOI: https://doi.org/10.1016/j.tree.2021.08.006
Bäurle I. Can't remember to forget you: chromatin-based priming of somatic stress responses. Semin Cell Dev Biol. 2018;83:133-9. Doi: 10.1016/j.semcdb.2017.09.032. DOI: https://doi.org/10.1016/j.semcdb.2017.09.032
Berger MMJ, Stammitti L, Carrillo N, Blancquaert E, Rubio B, Teyssier E, Gallusci P. Epigenetics: an innovative lever for grapevine breeding in times of climatic changes. OENO One. 2023;57(2):265-82. Doi: 10.20870/oeno-one.2023.57.2.7405. DOI: https://doi.org/10.20870/oeno-one.2023.57.2.7405
Boulias K, Greer EL. Means, mechanisms and consequences of adenine methylation in DNA. Nat Rev Genet. 2022;23(7):411-28. Doi: 10.1038/s41576-022-00456-x. DOI: https://doi.org/10.1038/s41576-022-00456-x
Crisp PA, Ganguly D, Eichten SR, Borevitz JO, Pogson BJ. Reconsidering plant memory: intersections between stress recovery, RNA turnover, and epigenetics. Sci Adv. 2016;2(2):e1501340. Doi: 10.1126/sciadv.1501340. DOI: https://doi.org/10.1126/sciadv.1501340
Delaunois B, Farace G, Jeandet P, Clément C, Baillieul F, Dorey S, Cordelier S. Elicitors as alternative strategy to pesticides in grapevine?: current knowledge on their mode of action from controlled conditions to vineyard. Environ Sci Pollut Res Int. 2014;21(7):4837-46. Doi: 10.1007/s11356-013-1841-4. DOI: https://doi.org/10.1007/s11356-013-1841-4
Ferrer M, Pereyra C, Salvarrey M, Arrillaga L, Fourment M. 'Tannat' (Vitis vinifera L.) as a model of responses to climate variability. Vitis. 2020;56:41-6. Doi: 10.5073/vitis.2020.59.41-46.
Fortes AM, Gallusci P. Plant stress responses and phenotypic plasticity in the epigenomics era: perspectives on the grapevine scenario, a model for perennial crop plants. Front Plant Sci. 2017;8:82. Doi: 10.3389/fpls.2017.00082. DOI: https://doi.org/10.3389/fpls.2017.00082
Fourment M, Ferrer M, Barbeau G, Quénol H. Local perceptions, vulnerability and adaptive responses to climate change and variability in a winegrowing region in Uruguay. Environ Manage. 2020;66(4):590-9. Doi: 10.1007/s00267-020-01330-4. DOI: https://doi.org/10.1007/s00267-020-01330-4
Fourment M, Ferrer M, Quénol H. Vitis vinifera L. cv. Tannat: respuesta a la variabilidad climática. Agrociencia (Uruguay). 2013;17(2):33-44. Doi: 10.31285/AGRO.17.433. DOI: https://doi.org/10.31285/AGRO.17.433
Fourment M, Piccardo P. What grapes and wines to expect with the drought? Agrocienc Urug. 2023;27(Supplement):e1206. Doi: 10.31285/AGRO.27.1206. DOI: https://doi.org/10.31285/AGRO.27.1206
Gallusci P, Agius DR, Moschou PN, Dobránszki J, Kaiserli E, Martinelli F. Deep inside the epigenetic memories of stressed plants. Trends Plant Sci. 2023;28(2):142-53. Doi: 10.1016/j.tplants.2022.09.004. DOI: https://doi.org/10.1016/j.tplants.2022.09.004
Gutierrez Gamboa G, Fourment M. Latin American viticulture adaptation to climate change: perspectives and challenges of viticulture facing up global warming. Cham: Springer; 2024. 250p. Doi: 10.1007/978-3-031-51325-1. DOI: https://doi.org/10.1007/978-3-031-51325-1
IPCC. Point of departure and key concepts. In: Climate Change 2022: impacts, adaptation and vulnerability: Working Group II Contribution to the Sixth Assessment Report of the Intergovernmental Panel on Climate Change. Cambridge: Cambridge University Press; 2023. p. 121-96. Doi: 10.1017/9781009325844.003. DOI: https://doi.org/10.1017/9781009325844.003
Latzel V, Rendina González AP, Rosenthal J. Epigenetic memory as a basis for intelligent behavior in clonal plants. Front Plant Sci. 2016;7:1354. Doi: 10.3389/fpls.2016.01354. DOI: https://doi.org/10.3389/fpls.2016.01354
Lauria M, Rossi V. Epigenetic control of gene regulation in plants. Biochim Biophys Acta. 2011;1809(8):369-78. Doi: 10.1016/j.bbagrm.2011.03.002. DOI: https://doi.org/10.1016/j.bbagrm.2011.03.002
Liu J, He Z. Small DNA methylation, big player in plant abiotic stress responses and memory. Front Plant Sci. 2020;11:595603. Doi: 10.3389/fpls.2020.595603. DOI: https://doi.org/10.3389/fpls.2020.595603
Lu AT, Xue L, Salfati EL, Chen BH, Ferrucci L, Levy D, Joehanes R, Murabito JM, Kiel DP, Tsai PC, Yet I, Bell JT, Mangino M, Tanaka T, McRae AF, Marioni RE, Visscher PM, Wray NR, Deary IJ, Levine ME, Quach A, Assimes T, Tsao PS, Absher D, Stewart JD, Li Y, Reiner AP, Hou L, Baccarelli AA, Whitsel EA, Aviv A, Cardona A, Day FR, Wareham NJ, Perry JRB, Ong KK, Raj K, Lunetta KL, Horvath S. GWAS of epigenetic aging rates in blood reveals a critical role for TERT. Nat Commun. 2018;9(1):387. Doi: 10.1038/s41467-017-02697-5. DOI: https://doi.org/10.1038/s41467-017-02697-5
Marfil C, Ibañez V, Alonso R, Varela A, Bottini R, Masuelli R, Fontana A, Berli F. Changes in grapevine DNA methylation and polyphenols content induced by solar ultraviolet-B radiation, water deficit and abscisic acid spray treatments. Plant Physiol Biochem. 2019;135:287-94. Doi: 10.1016/j.plaphy.2018.12.021. DOI: https://doi.org/10.1016/j.plaphy.2018.12.021
Mira de Orduña R. Climate change associated effects on grape and wine quality and production. Food research international. 2010;43(7):1844-55. DOI: https://doi.org/10.1016/j.foodres.2010.05.001
Mozgova I, Mikulski P, Pecinka A, Farrona S. Epigenetic mechanisms of abiotic stress response and memory in plants. In: Alvarez-Venegas R, De-la-Peña C, Casas-Mollano JA (Eds.). Epigenetics in plants of agronomic importance: fundamentals and applications: transcriptional regulation and chromatin remodelling in plants. Cham: Springer; 2019. p. 1-64. DOI: https://doi.org/10.1007/978-3-030-14760-0_1
Niederhuth CE, Bewick AJ, Ji L, Alabady MS, Kim KD, Li Q, Rohr NA, Rambani A, Burke JM, Udall JA, Egesi C, Schmutz J, Grimwood J, Jackson SA, Springer NM, Schmitz RJ. Widespread natural variation of DNA methylation within angiosperms. Genome Biol. 2016;17(1):194. Doi: 10.1186/s13059-016-1059-0. DOI: https://doi.org/10.1186/s13059-016-1059-0
Pikaard CS, Mittelsten Scheid O. Epigenetic regulation in plants. Cold Spring Harb Perspect Biol. 2014;6(12):a019315. Doi: 10.1101/cshperspect.a019315. DOI: https://doi.org/10.1101/cshperspect.a019315
Rubio B, Stammitti L, Cookson SJ, Teyssier E, Gallusci P. Small RNA populations reflect the complex dialogue established between heterograft partners in grapevine. Hortic Res. 2022;9:uhab067. Doi: 10.1093/hr/uhab067. DOI: https://doi.org/10.1093/hr/uhab067
Shangguan L, Fang X, Jia H, Chen M, Zhang K, Fang J. Characterization of DNA methylation variations during fruit development and ripening of Vitis vinifera (cv. 'Fujiminori'). Physiol Mol Biol Plants. 2020;26(4):617-37. Doi: 10.1007/s12298-020-00759-5. DOI: https://doi.org/10.1007/s12298-020-00759-5
Sun X, Fan G, Su L, Wang W, Liang Z, Li S, Xin H. Identification of cold-inducible microRNAs in grapevine. Front Plant Sci. 2015;6:595. Doi: 10.3389/fpls.2015.00595. DOI: https://doi.org/10.3389/fpls.2015.00595
Tan JW, Rodríguez López CM. Epigenomics: a new tool for the generation of climate resilient grapevines. Front Hortic. 2023;2. Doi: 10.3389/fhort.2023.1116866. DOI: https://doi.org/10.3389/fhort.2023.1116866
Tang D, Gallusci P, Lang Z. Fruit development and epigenetic modifications. New Phytol. 2020;228(3):839-44. Doi: 10.1111/nph.16724. DOI: https://doi.org/10.1111/nph.16724
Varela A, Ibañez VN, Alonso R, Zavallo D, Asurmendi S, Gomez Talquenca S, Marfil CF, Berli FJ. Vineyard environments influence Malbec grapevine phenotypic traits and DNA methylation patterns in a clone-dependent way. Plant Cell Rep. 2021;40(1):111-25. Doi: 10.1007/s00299-020-02617-w DOI: https://doi.org/10.1007/s00299-020-02617-w
Venios X, Gkizi D, Nisiotou A, Korkas E, Tjamos SE, Zamioudis C, Banilas G. Emerging roles of epigenetics in grapevine and winegrowing. Plants (Basel). 2024;13(4):515. Doi: 10.3390/plants13040515. DOI: https://doi.org/10.3390/plants13040515
Wibowo A, Becker C, Durr J, Price J, Spaepen S, Hilton S, Putra H, Papareddy R, Saintain Q, Harvey S, Bending GD, Schulze-Lefert P, Weigel D, Gutierrez-Marcos J. Partial maintenance of organ-specific epigenetic marks during plant asexual reproduction leads to heritable phenotypic variation. Proc Natl Acad Sci USA. 2018;115(39):E9145-E9152. Doi: 10.1073/pnas.1805371115. DOI: https://doi.org/10.1073/pnas.1805371115
Xie H, Konate M, Sai N, Tesfamicael KG, Cavagnaro T, Gilliham M, Breen J, Metcalfe A, Stephen JR, De Bei R, Collins C, Lopez CMR. Global DNA Methylation Patterns Can Play a Role in Defining Terroir in Grapevine (Vitis vinifera cv. Shiraz). Front Plant Sci. 2017;8:1860. Doi: 10.3389/fpls.2017.01860. DOI: https://doi.org/10.3389/fpls.2017.01860
Zemach A, Kim MY, Hsieh PH, Coleman-Derr D, Eshed-Williams L, Thao K, Harmer SL, Zilberman D. The Arabidopsis nucleosome remodeler DDM1 allows DNA methyltransferases to access H1-containing heterochromatin. Cell. 2013;153(1):193-205. Doi: 10.1016/j.cell.2013.02.033. DOI: https://doi.org/10.1016/j.cell.2013.02.033
Zhang YY, Latzel V, Fischer M, Bossdorf O. Understanding the evolutionary potential of epigenetic variation: a comparison of heritable phenotypic variation in epiRILs, RILs, and natural ecotypes of Arabidopsis thaliana. Heredity (Edinb). 2018;121(3):257-65. Doi: 10.1038/s41437-018-0095-9. DOI: https://doi.org/10.1038/s41437-018-0095-9
Zhu Z, Li Q, Gichuki DK, Hou Y, Liu Y, Zhou H, Xu C, Fang L, Gong L, Zheng B, Duan W, Fan P, Wang Q, Xin H. Genome-wide profiling of histone H3 lysine 27 trimethylation and its modification in response to chilling stress in grapevine leaves. Hortic Plant J. 2023;9(3):496-508. Doi: 10.1016/j.hpj.2023.03.002. DOI: https://doi.org/10.1016/j.hpj.2023.03.002
Downloads
Publicado
Como Citar
Edição
Seção
Licença
Copyright (c) 2024 Agrociencia Uruguay

Este trabalho está licenciado sob uma licença Creative Commons Attribution 4.0 International License.
| Métricas do artigo | |
|---|---|
| Vistas abstratas | |
| Visualizações da cozinha | |
| Visualizações de PDF | |
| Visualizações em HTML | |
| Outras visualizações | |